Of special interest is the question, whether proteins and nucleic acids could arise.
S. W. FOX (University of Miami) studied the formation of proteins from amino acids from the 1960th on. His studies show that proteins and / or protein-like polymers (so-called proteinoids) develop when heating a mixture of amino acids up to 150 – 200 oC. The incubation time takes several days. The yield is higher when adding polyphosphate, polymerization then occurs already at 70 oC. These polymers display two decisive features:
Proteinoids show catalytic activity. Their range of properties does not cover all the features of modern enzymes, especially not their reaction velocity, but proteinoids are obviously well-suited to be perfected by evolution. The kinetic of some reactions shows, that proteinoids do already display a co-operative (allosteric) behaviour.
Proteinoids form structures. They have the tendency to aggregate (development of a quaternary structure). They form hollow spheres (so-called microspheres) in aqueous solutions, an important step towards the evolution of the cell. Microspheres may be regarded as a precursor of protocells. They share one important feature with cells: they form a compartment that separates a reaction from the surrounding.
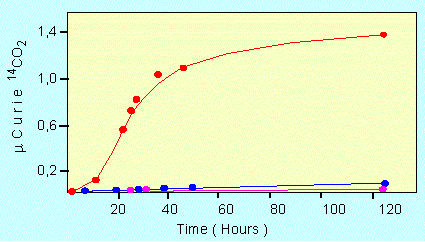
The catalytic activity of a proteinoid. The observed reaction was the decarboxylation of pyruvate, red: in the presence of a proteinoid, blue: in the presence of a mixture of free amino acids, and pink: without additive (according to S. W. FOX and K. DOSE, 1972)
It seems thus as if self-assembly would be an inherent feature of matter. It is caused by the tendency of hydrophobic parts of molecules to form the maximal number of weak interactions excluding water. The analysis of the assembly of proteinoids made clear that polymerization does not follow the laws of chance alone. The amino acid composition of proteinoids differs considerably from that of the amino acid mixture, i.e. the probability to be incorporated into a proteinoid is not the same for all amino acids, but certain amino acids like aspartic acid are preferred. The physico-chemical properties of monomers as well as the starting sequence of a newly developing polymer determine, which further amino acids will be incorporated into the growing polypeptide chain. The aim is always to acquire a state of low energy that is characterized by two criteria:
Such preferences are not unusual at all from a physico-chemical point of view, since in crystal formation, too, similar molecules or ions or such that fit together due to structure or charge, i.e. complementary molecules or ions assemble also excluding other, non-complementary molecules or ions in this process.
Microspheres may gain size by incorporating additional proteinoids, but become hardly ever larger than ~15 µm in diameter. After reaching this value, they do either divide or bud off. Small microspheres are able to fuse with each other. They are very stable in aqueous solutions, but their stability decreases and the ability to distort increases considerably as soon as lipid molecules are incorporated. Lipids, too, may have occurred during the abiotic environment of the earth surface.
Studies of cross-sections of microspheres with electron microscopes show that proteinoids are organized in membrane-like double layers, though the dimensions of these layers does not correspond with that of today’s double membrane.
Conclusively, it can be stated that cell-like structures were able to evolve from proteinoids with the aid of other molecules like ions, lipids, etc, and that a number of biochemical processes were catalyzed at their surfaces and inside of them. Moreover, the membrane-like outer layer became selective for certain molecules while at the same time excluding others (a state called selective permeability). Fusion of microspheres of different compositions and functions could have resulted in fusion products performing the reactions of all of their parental microspheres. At the same time, they show, that the occurrence, stability, and the performance of the microspheres were subject to selection.
Do these structures constitute a living system? The answer is clearly no, even though they display a number of features typical for living cells: catalysis, allosteric behaviour, self-assembly, cell-like appearance, membrane-like layers, the ability to divide and to fuse, selective permeability, and selection. They do nevertheless lack the ability of identical duplication, i.e. the ability to develop. This lack is caused by the inability of proteins to store and pass on the information they contain. Imagine, nevertheless, that a number of catalytic proteinoid entities would join together and catalyze the production of each other. The reaction partners would then be elements of a cycle that could, as a whole, replicate. The ability to replicate would accordingly be a property of the system, not that of the single components. But as a consequence, the system would be intolerant against changes like mutations, since a mutation of one of the cycle’s components would necessarily lead to either an impairment or a disruption of the cycle causing a lesser efficiency or a break-down of the system. The system would thus become extinct. A mutation of such an imaginary system had virtually no change to manifest as it would have to initiate a completely new cycle in order to do so. The probability of such an event is enormously small. A positive effect of a mutation in such a system could be regarded not as evolution, but as a revolution, where the whole information collected before the mutation occurred, i.e. the order
A > B > C >........ X > A
would at once be lost and replaced by something radically new. M. EIGEN (Max-Plank Institute of Biophysical Chemistry, Göttingen, Germany) calculated the coming into being of such cycles and their probability to change and found that both the age and the size of the total universe would not be sufficient to develop a successful strategy based upon them.
How did evolution solve this dilemma? Is the evolution of life possible on the basis of another type of molecules ?
In contrast to proteinoids and proteins, nucleic acids have matrix functions and are thus able to replicate identically, the necessary prerequisite to carry genetic information. Under abiotic conditions, nucleotides can polymerize forming polynucleotides. Extensive series of experiments performed in the lab of L. E. ORGEL showed, that phosphodiester bonds form between nucleotides after addition of carbodiimide. The resulting bonds were not just of today’s 3’ >5’ type, but also 2’>5’ and 5’>5’ types. Nucleic acids have the tendency to form base pairs as a consequence of the fact that purine and pyrimidine bases are complementary to each other. The base pairs occur either between two single strands or between sections of the same strand. Single and double nucleotide strands form statistically in aqueous solutions, no definite tertiary structures are formed. tRNA seems to be an exception, but its tertiary structure is based on the enzyme-aided modification of single bases.
See e.g.
Double strands resist thermodynamic denaturation, i.e. denaturation caused by temperature-dependent molecular movements better than single strands. They are more stable, a selective advantage. The longer a polynucleotide strand is, the more stable it is, too. This is on one hand caused by the number of hydrogen bonds between opposing bases (two in the case of A = U/T pairs, three with G = C pairs), on the other hand on the so-called stacking energy between neighbouring base pairs.
The ability to form hydrogen bonds is not restricted to the mentioned base pairs alone, two hydrogen bonds each are also formed between U = U, T = T, A = A, G = G and G = T pairs, for example. Why then do only the A = T/U and G = C pairs occur in all today’s known nucleic acids, and why are they exclusively linked via 3’ > 5’ phosphodiester bonds?
The answer is, that polymers where each purine opposes a pyrimidine and where the monomers are connected by always the same type of bond form regular structures. Regularity and the resulting complementarity again are the basis of a high stability.
In contrast to RNA, DNA occurs usually as WATSON-CRICK double helices. There do also exist RNA double strands and RNA that is partially double-stranded, but its molecular configuration is different. In contrast to the WATSON – CRICK model are the base pairs not arranged vertically to the axis. The consequence is a measurably lower stability against thermal denaturation and that seems to be the reason why its is DNA and not RNA that became the carrier of the organisms’ genetic information. This is another clue for selection not being a biological phenomenon, but a basic feature of matter.
Why did nucleic acids become the carriers of genetic information? Besides their higher stability, they are also able to replicate identically due to their ability to function as a matrix, and they offer a supply of several bases (A, T, C, G) that can be arranged in any sequence thus composing theoretically unlimited amounts of information units. Polynucleotide double strands consisting of only C and G are more stable than those containing additionally A and T, but their content of information would be reduced drastically.
A sequence of bases per se cannot be regarded as an information, because in order to become information, it is necessary that somebody or something is able to decode it. The matrix properties are thus a prerequisite for the sequence of bases to become information. Nucleic acids are consequently self-instructing information carriers.
The incorporation of ‘wrong’ base pairs during replication leads to mistakes. If such a mistake has occurred, it cannot be corrected any more. It will thus effect all following cycles of replication and will be passed on to all subsequent generations of molecules. In such a case, the sequence of bases would be erroneous, and – in the case that further mistakes occur - it could even be lost completely or be replaced successively by another sequence. The ability to replicate would nevertheless remain.
Would such a system be able to go through evolution? May life have evolved on a system of nucleic acids alone? The answer is no.
Nucleic acids lack a control mechanism for the maintenance and the safeguarding of the information contained in the sequence of bases. It lacks thus also the ability to maintain valuable information and to distinguish between valuable and worthless information. How can the information contained in the base sequences have a value at all? And how is valuable genetic information linked to living systems?
|
|