- Eukaryotic Gene Structure
- The eukaryotic genome contains a significant amount of DNA that is neither transcribed nor translated.
- Prokaryotic genomes do not contain much DNA that is not used either for direct or indirect information content.
- In some cases, prokaryotic genes may overlap such as in one case the sequence ....UGAUG... where the UGA is the stop codon for one gene and the AUG is the initiation codon for the next gene.
- There are instances of a sequence of DNA being read in different directions as functional parts of separate genes.
- Eukaryotic genes are usually surrounded by non-transcribed DNA although some of this DNA may contain regulatory information.
- The large majority of eukaryotic genes contain blocks of DNA that are noncoding and do not end up in the mature mRNA. This is in direct contrast to prokaryotic genes, in general, have a direct base to base correspondence between the gene and the mRNA.
- the DNA sequences that contain the information for amino acid sequences and end up in the mature mRNA are called exons for expressed sequences
- the DNA sequences that are noncoding and do not end up in the mature mRNA are called introns for interveing sequences.
- Eukaryotic genes usually occur as solitary genes on a chromosome as opposed to the closely linked genes of bacterial operons. Genes associated with different functions in the same biochemical pathway are often located on different chromosomes.
- the gene coding for the enzyme that creates the precursor for the A,B red blood cell antigen is located on chromosome #19 (fucosyltransferase 1)
- the gene with the alleles for creating either the A or B antigen is located on chromosome #9 (glycosyltransferase)
- Eukaryotic genes often exist as members of a"gene family". That is, there may be more than one copy of a similar or nearly identical DNA sequence in the genome.
- the globin genes comprise a gene family
- the myoglobin gene is located on chromosome 22q
- the
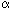 -globin gene is located on chromosome 16p
-globin gene is located on chromosome 16p
- the
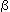 -globin gene is located on chromsome 11p
-globin gene is located on chromsome 11p
- some of the members of a gene family may be non-functional because mutations render them so - these are known as"pseudogenes"
- Eukaryotic Gene Transcripts Must be Processed
- The initial transcript from a eukaryotic gene is processed in three ways before it can leave the nucleus to participate in the translation process.
- The 5' end of the RNA transcript is modified while the RNA is being produced
- The 5' end of the RNA transcript is a purine triphosphate.
- An enzyme, guanylyl transferase, attaches a guanosine to the 5' end of the transcript. The enzyme is associated with RNA pol II which only produces mRNA.
- The linkage is made using the 5' carbon of the ribose attached to the guanosine.
- In effect, the 5' end of the RNA becomes a 3'OH of the added guanosine.
- This added base is called a 5' cap and marks the RNA as being a mRNA.
- The 3' end of the mRNA transcript is modified by the addition of a poly A sequence
- Beyond the 3' end of the coding regions of each mRNA transcript is the sequence (or one very similar) ...AAUAAA... and another sequence further along ..GUUU..
- This sequence is recognized by an enzyme which cleaves the RNA a short distance away in the 3' direction from the AAUAAA site(downstream).
- Another enzyme set adds a number of adenines to the end of the RNA producing a poly A tail on the 3' end of the mRNA.
- mRNA transcripts lacking the polyA addition are degraded rapidly in the nucleus.
- The fact that mRNAs have a polyA 3"tail" was utilized in the isolation of mRNAs by binding them to sequences of polyT.
- The noncoding sequences in the RNA are removed by a"splicing mechanism"
- The 5' and 3' ends of intron contain invariant sequences (GU....AG).
- For many RNAs, the process of splicing is carried out by small nuclear ribonucleoprotein particles (snRNPs) that consist of proteins and an RNA molecule (one of those"other").
- Some of the splicing depends on base pairing between the RNA of the snRNP and the mRNA.
- It is estimated that over 100 proteins are involved in RNA splicing.
- The result of splicing is the bringing together of contiguous coding sequences to produce a mature mRNA.
- The exons are often associated with specific functional domains of the protein.
- This suggests that genes may evolve by the"shuffling" of exons which provides a way to put together new combinations of functional domains.
- Splice site mutations can lead to errors in splicing.
- destroy existing splice site and produce missense
- create new splice site in intron to produce two splices - one good and one bad
- the disease lupus erythromatosus is accompanied by the production of antibodies to a snRNP
- examples
- Transcriptional Control of Gene Expression
- Eukaryotic genes are not regulated in the same manner as prokaryotic genes. There are many more protein factors involved and both positive and negative regulation are the rule rather than the alternatives.
- The transcription start is usually associated with an AT rich DNA sequence about 25 bp away from the initiation - the TATA box
- Upstream anywhere from 50 to 100 bases from the TATA box is another sequence associated with transcription initiation - the CAAT box. This sequence may not be necessary in all genes.
- In the general vicinity, there may be other sequences know as enhancer sequences. These sequences may be 100 to 200 base pairs in size and up to 50 kilobases away from the affected gene.
- Transcription is not initiated by the simple binding of the RNA pol II with the CAAT and TATA boxes.
- It requires the binding of a protein transcription factor and the binding of other proteins. And some of the proteins are activated by phosphorylation.
- In addition, there are repressor proteins that can bind to the initiation complex to interfere with transcription initiation.
- Some of the complexity of regulation allows for tissue specificity or temporal specificity.
- some genes are expressed in widely different tissues
- but the regulatory elements that function for expression in one tissue are not required for expression in another tissue
- Some regulation is less precise and involves"shutting down" a portion or most of a chromosome
- X-inactivation involves the random inactivation of most of the genes on one of the two X chromosomes in a female early in development.
- At an early stage of embryonic development in individuals with more than one X
chromosome per cell, the extra X chromosome(s) condense, become heterochromatic, and
are the genes are no longer transcribed.
- In mammals, the X chromosome undergoing the inactivation event is a random choice
from one cell to another.
- Once an X chromosome has been inactivated, it remains inactivated in the daughter cells derived from the original cell.
- This results in a"mosaic effect" or a"patchwork effect" in the individual and can be demonstrated in humans and other mammals.
- cats
- humans
- X-inactivation can be demonstrated at the cellular level
- Remembering that XO individuals are shorter than one would expect and XXY and XYY individuals are taller than one would expect, the prediction is that not all of the genes on an inactive X are inactive.
- That seems to be case as illustrated by certain enzyme coding genes.
- The addition of methyl groups to DNA bases is associated with reduced transcriptional activity
- For some genes, regulation involves moving the gene to a new position.
- For other genes, there is a selective synthesis of the DNA containing those genes so that the number of gene copies is increased tremendously. This gene amplification occurs in frogs and fish for the rRNA genes and is necessasry in order for the eggs to have enough rRNA produced.
- Post-Transcriptional Control of Gene Expression
- Some messages undergo alternate splicing depending on what tissue they are located in. The regulation is at the level of snRNP production.
- The stability of a class of mRNA can be controlled.
- Some short-lived mRNAs have multiple copies of the sequence AUUUA which may act as a target for degradation.
- the hormone prolactin enhances the stability of the mRNA for the milk protein casein
- high levels of iron decrease the stability of the mRNA for the receptor that brings iron into cells
- Translational Control of Gene Expression
- Protein factors may enhance the reading of particular mRNAs such as excess heme leads to increased reading of globin mRNA in RBC.
- The 5' cap on a mRNA is usually modified by the addition of a methyl group to the guanine. mRNAs with a cap that has not been modified are not translated but are stored as"masked messengers". Translation is initiated when the cap is modified.
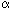 -globin gene is located on chromosome 16p
-globin gene is located on chromosome 16p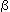 -globin gene is located on chromsome 11p
-globin gene is located on chromsome 11p