The data of W. BATESON and
colleagues on Lathyrus odoratus provided an experimental
approach to the localization of genes on chromosomes. In Lathyrus
odoratus , two characteristics are coupled that are nevertheless
sometimes separated from each other. BATESON's results were not
sufficient to draw further conclusions. It was data gained on
Drosophila that allowed more insight into the way genes are
organized on chromosomes.
The chromosome theory claims that the number of
linkage groups corresponds to the
number of chromosomes in a haploid set (n= 1). But actually, at first
in Pisum sativum later on also in other species, more linkage
groups than n = 1 chromosomes were detected. H.
de VRIES assumed therefore in 1903 that the surplus in the number
of linkage groups was founded on the exchange of fragments between
chromosomes. The necessary structural preconditions are given during
the prophase of meiosis when the homologous chromosomes pair. A. F.
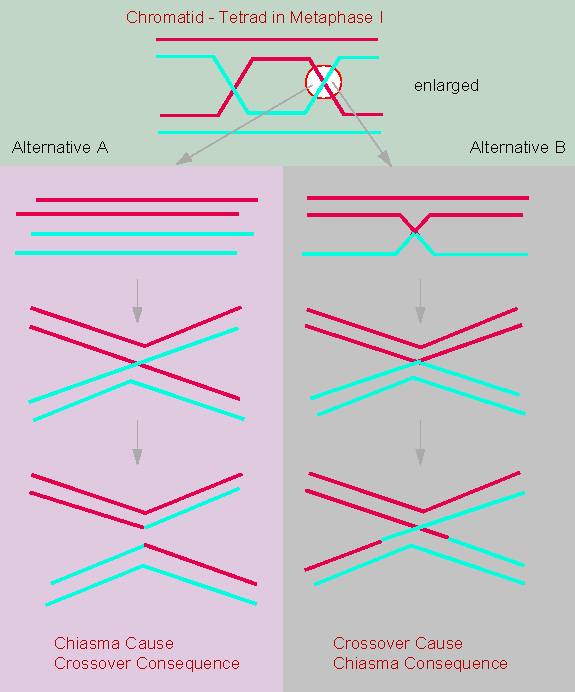
JANSSENS observed the cross of two chromosomal arms during the
diplotene of meiosis in
Batrachoseps attenuatus, an amphibian, and called this
phenomenon chiasma. He, too, saw
the possibility that the chromatids might break in such a position
and that the broken parts might join the 'wrong' chromatid, thus
causing an exchange of fragments.
It was T. H. MORGAN who could finally show that an exchange of the hereditary factors between two X-chromosomal characteristics occurs and that it is due to a crossing-over. In other words: genetic analyses lead to the assumption of an exchange of chromosomal fragments.
The relative probability of crossing-overs is decisive for
the drawing-up of a genetic map. If two loci of
one chromosome are in vicinity, then the probability that they are
separated by a crossing over is low. It increases with growing
distance. A. H. STURTEVANT analyzed the exchange frequency of several
X-chromosomal genes this way. He could prove that it is indeed
correlated to the distances between the genes and that these
distances are additive. That means that the distance between gene A
and gene C is the same as the sum of the distances between gene A and
gene B and between gene B and gene C. This again indicates that the
gene loci A, B and C are arranged on the chromosome one behind the
other. The method has its weaknesses, since the actual values for
large gene distances are often smaller than calculated. This is
caused by a phenomenon called interference that happens, because the
preceding crossing-over has an influence on the probability of the
following ones.
The first, at the beginning still very simple gene map of a linkage group was that of the X-chromosome of Drosophila. It was soon followed by additions and further maps. The first gene map of a plant chromosome that consisted of only three loci was drawn up as soon as 1926 (L. J. STADLER, University of Missouri, Columbia, 1926).
After the mapping of numerous new gene loci, the distances originally determined by STADLER had to be stated more precisely, since they were falsified by double crossing-overs. It was the precise analysis done with the help of numerous gene loci, so called marker genes that led to an approach to the real distances.
Apart from meiotic crossing-overs, a mitotic crossing-over with sister chromosome exchange is known, too.
|
|